
|
|
|
|
| DTI-Query 1.1 | CINCH 1.0 BETA |
There are a variety of tools available for estimating tensors from raw diffusion-weighted images (AFNI, FSL, etc.) We have partnered with the AFNI group at NIH to make it easy to use DTI-Query/CINCH with AFNI. See here for instructions on converting AFNI data into the format needed by DTI-Query.
Regardless of the software you use for this step, you will need to produce a single NIFTI output file containing the tensor coefficients. This file stores six volumes, one volume for each coefficient of the tensor data. The order of the volumes may be arbitrary, and can be specified as a parameter to the DTI-Query/CINCH precomputation interface. (By default, we expect Dxx, Dxy, Dyy, Dxz, Dyz, Dzz, as per the NIFTI conventions.) The input NIFTI image can be either 4D or 5D (recommended). In the case of a 4D file, the tensor component is indexed by the fourth dimension. In the case of a 5D file, the tensor component is indexed by the fifth dimension (NIFTI expects the fourth dimension to be time).
Optionally, you can compute a NIFTI mask image for use in tractography; dtiprecompute will only seed or trace pathways in regions where the voxel values are greater than zero. This allows you to avoid tracing pathways through regions of the brain where you know that pathways do not exist. You can use AFNI to automatically produce such a mask volume.
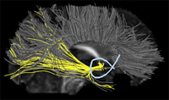
Next, you need to compute pathways from the outputs of steps 1 and 2. Use the dtiprecompute_cmd (command line tool) or dtiprecompute_gui (GUI version) to do this. The input parameters are described here. The outputs include a pathway database file (.pdb) and an FA (Fractional Anisotropy) map (.nii.gz).
Optionally, you can compute a matrix of inter-pathway distances, using the dmatrix program included in the distribution. CINCH uses this information to enable interactive growing and shrinking of pathway selections. CINCH will still work without this step, but you will not be able to grow or shrink selections.
The command-line interface to dmatrix is simple:
dmatrix /p yourpathways.pdb
It produces a file called "yourpathways.pdb.dis" that contains the distance matrix. CINCH will automatically read this file if it is present, enabling the growing and shrinking of pathway selections.
(Warning: This step can take considerable time to run - it is O(n^2) in the number of pathways.)
Questions? Problems? E-mail us.