|
|
-
Zahid Hossain, Engin W. Bumbacher Alison Brauneis, Monica Diaz, Andy Saltarelli, Paulo Blikstein, Ingmar H Riedel-Kruse "Design Guidelines and Empirical Case Study for Scaling Authentic Inquiry-based Science Learning via Open Online Courses and Interactive Biology Cloud Labs," International Journal of Artificial Intelligence in Education (2017): 1-30.
Abstract:
[IJAIED]
The Next Generation Science Standards (NGSS) and other national frameworks are calling for much more sophisticated approaches to STEM education, centered around the integration of complex experimentation (including real labs, not just simulations), data collection and analysis, modeling, and data-driven argumentation, i.e., students can behave like real scientists. How to implement such complex approaches in scalable ways is an unsolved challenge - both for presential and distance education. Here we report on the iterative design and large-scale deployment of an open online course with a "biology cloud experimentation lab" (using living cells) that engaged remote learners (> 300 students) in the scientific practices of experimentation, modeling and data analysis to investigate the phototaxis of a microorganism. We demonstrate (1) the robustness and scalability of the cloud lab technology (> 2,300 experiments run), (2) the design principles and synergistic integration of multiple UI and learning activities and suitable data formats to facilitate NGSS-aligned science activities, and (3) design features that leverages the natural variability of real biology experiments to instigate authentic inquiry. This platform and course content are now suited for large-scale adaptation in formal K-16 education; and we provide recommendations for inquiry-based science learning in general.
|
|
|
|
-
Amy T Lam , Karina Giselle Samuel-Gama , Jonathan Caleb Griffin , Matthew Austin Loeun , Lukas C Gerber , Zahid Hossain , Nate J. Cira , Seung Ah Lee and Ingmar Riedel-Kruse, "Device and Programming Abstractions for Spatiotemporal Control of Active Micro-particle Swarms," Lab on a Chip, 2017.
Abstract:
[Lab On a Chip]
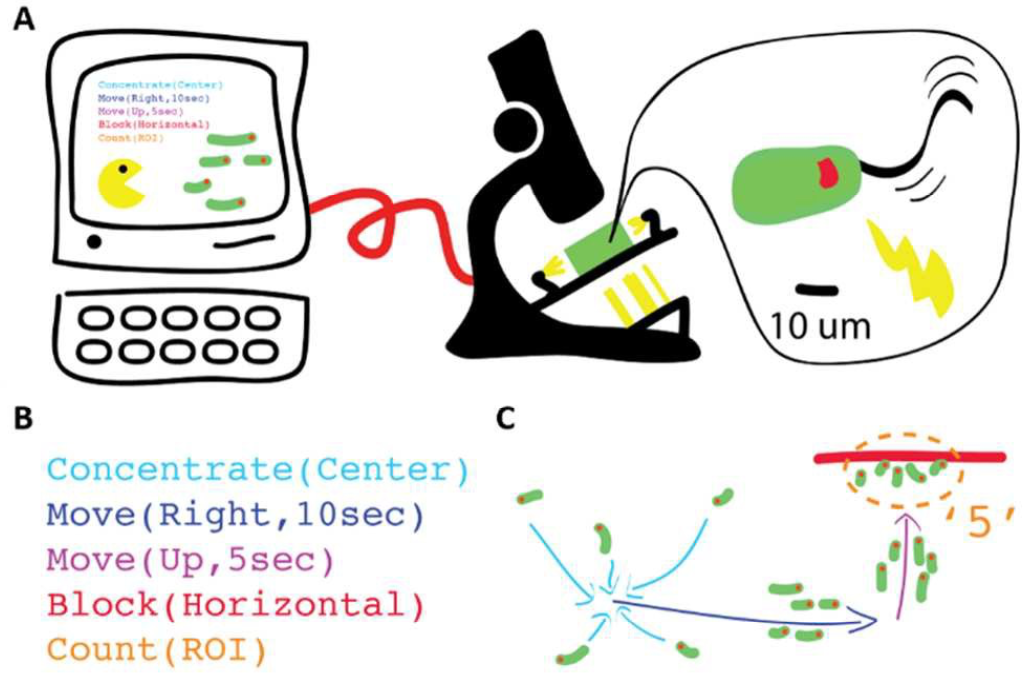
We present a hardware setup and a set of executable commands for spatiotemporal
programming and interactive control of a swarm of self-propelled microscopic agents
inside a microfluidic chip. In particular, local and global spatiotemporal
light stimuli are used to direct the motion of ensembles of Euglena gracilis,
a unicellular phototactic organism. We develop three levels of programming abstractions
(stimulus space, swarm space, and system space) to create a scripting language for
directing swarms. We then implement a multi-level proof-of-concept biotic game using
these commands to demonstrate their utility. These device and programming concepts will
enhance our capabilities for manipulating natural and synthetic swarms, with future
applications for on-chip processing, diagnostics, education, and research on collective behaviors.
|
|
|
|
-
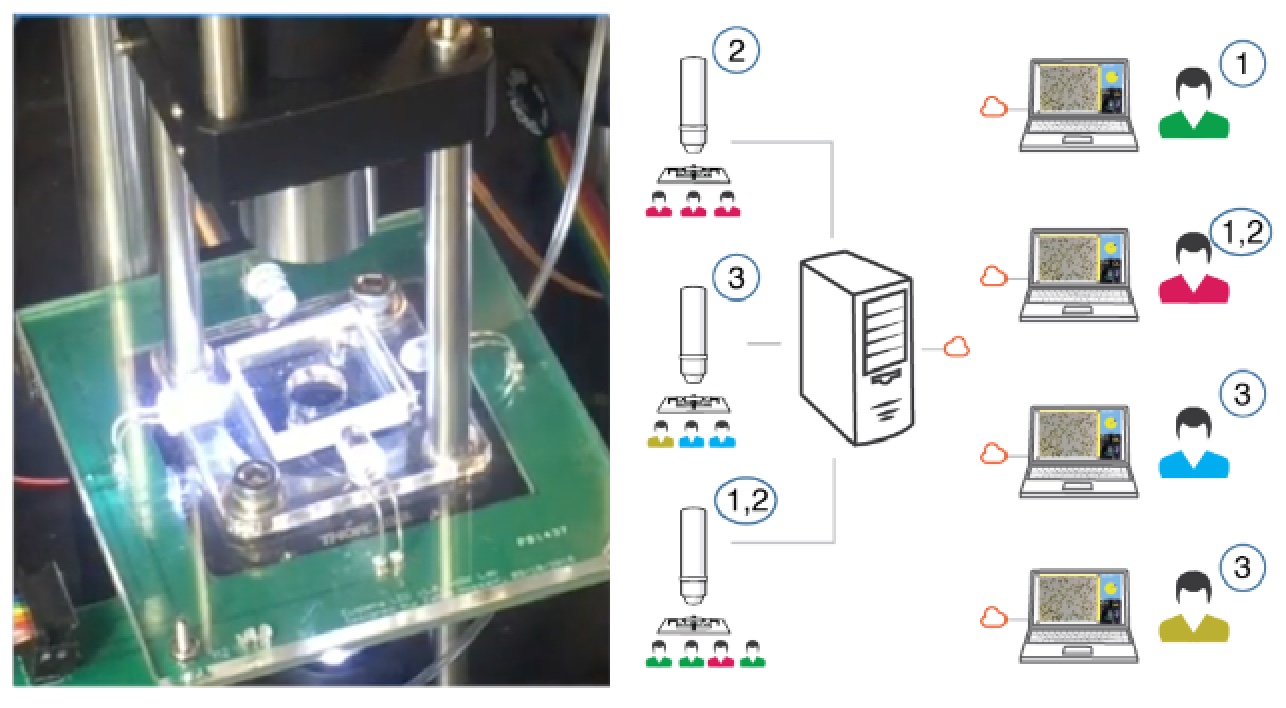
Zahid Hossain, Engin W. Bumbacher, Alice M. Chung, Honesty Kim, Casey Litton, Ashley D. Walter, Sachin N. Pradhan, Kemi Jona, Paulo Blikstein, Ingmar H. Riedel-Kruse, "Interactive and Scalable Biology Cloud Experimentation for Scientific Inquiry and Education," Nature Biotechnology, 34(12):1293-1298, 12 2016.
Abstract:
[Nature Biotech] [Youtube] [Stanford News]
A real-time interactive, fully automated, low-cost and scalable biology cloud experimentation platform could provide access to scientific experimentation for learners and researchers alike.
|
|
|
|
-
Dominik L. Michels, Dmitry A. Lyakhov, Vladimir P. Gerdt, Zahid Hossain, Ingmar H. Riedel-Kruse, Andreas G. Weber , On the General Analytical Solution of the Kinematic Cosserat Equations,
in the Proceedings of Computer Algebra in Scientific Computing CASC, 2016
Abstract:
[PDF (Authors' version)] [Project page]
Based on a Lie symmetry analysis, we construct a closed form solution to the kinematic part of the (partial differential) Cosserat equations describing the mechanical behavior of elastic rods. The solu- tion depends on two arbitrary analytical vector functions and is analyt- ical everywhere except a certain domain of the independent variables in which one of the arbitrary vector functions satisfies a simple explicitly given algebraic relation. As our main theoretical result, in addition to the construction of the solution, we proof its generality. Based on this observation, a hybrid semi-analytical solver for highly viscous two-way coupled fluid-rod problems is developed which allows for the interactive high-fidelity simulations of flagellated microswimmers as a result of a substantial reduction of the numerical stiffness.
|

|
|
|
-
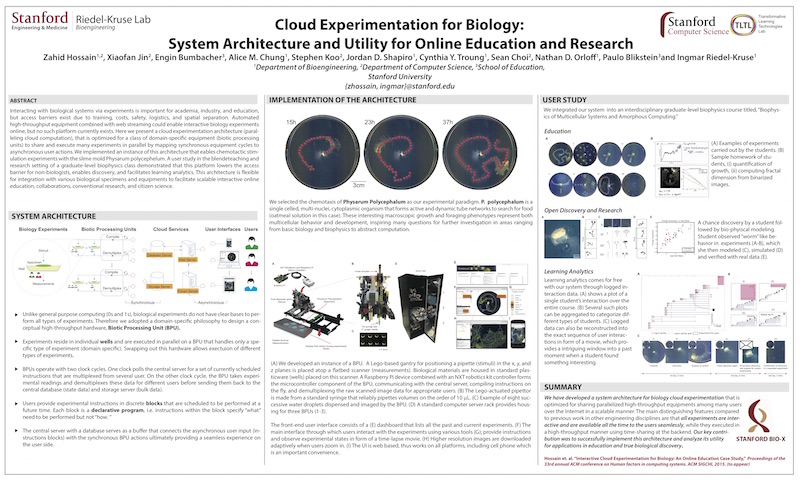
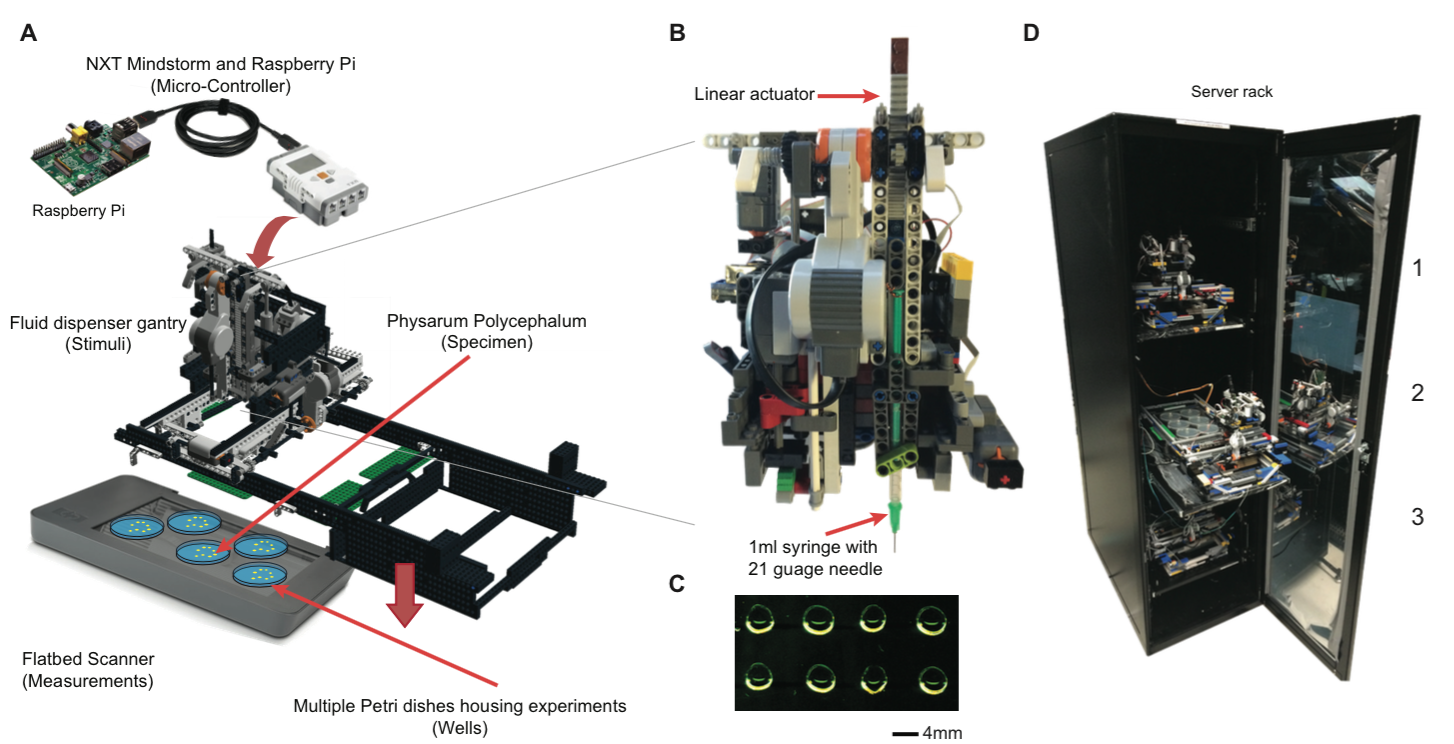
Zahid Hossain, Xiaofan Jin, Engin W. Bumbacher, Alice M. Chung, Stephen Koo, Jordan D. Shapiro, Cynthia Y. Truong, Sean Choi, Nathan D. Orloff, Paulo Blikstein, Ingmar H. Riedel-Kruse, "Interactive Cloud Experimentation for Biology: An Online Education Case Study," (Honorable Mention) Proceedings of the 33rd Annual ACM Conference on Human Factors in Computing Systems, SIGCHI, (3681-3690), 2015.
Abstract: [PDF] [Supplementary Materials]
[Slides - Keynote Format]
[Youtube]
[Stanford News]
Interacting with biological systems via experiments is important
for academia, industry, and education, but access barriers
exist due to training, costs, safety, logistics, and spatial
separation. High-throughput equipment combined with web
streaming could enable interactive biology experiments online,
but no such platform currently exists. We present a cloud
experimentation architecture (paralleling cloud computation),
which is optimized for a class of domain-specific equipments
(biotic processing units - BPU) to share and execute many
experiments in parallel remotely and interactively at all time.
We implemented an instance of this architecture that enables
chemotactic experiments with a slime mold Physarum Polycephelum.
A user study in the blended teaching and research
setting of a graduate-level biophysics class demonstrated that
this platform lowers the access barrier for non-biologists, enables
discovery, and facilitates learning analytics. This architecture
is flexible for integration with various biological specimens
and equipments to facilitate scalable interactive online
education, collaborations, research, and citizen science.
|
|
|
|
-
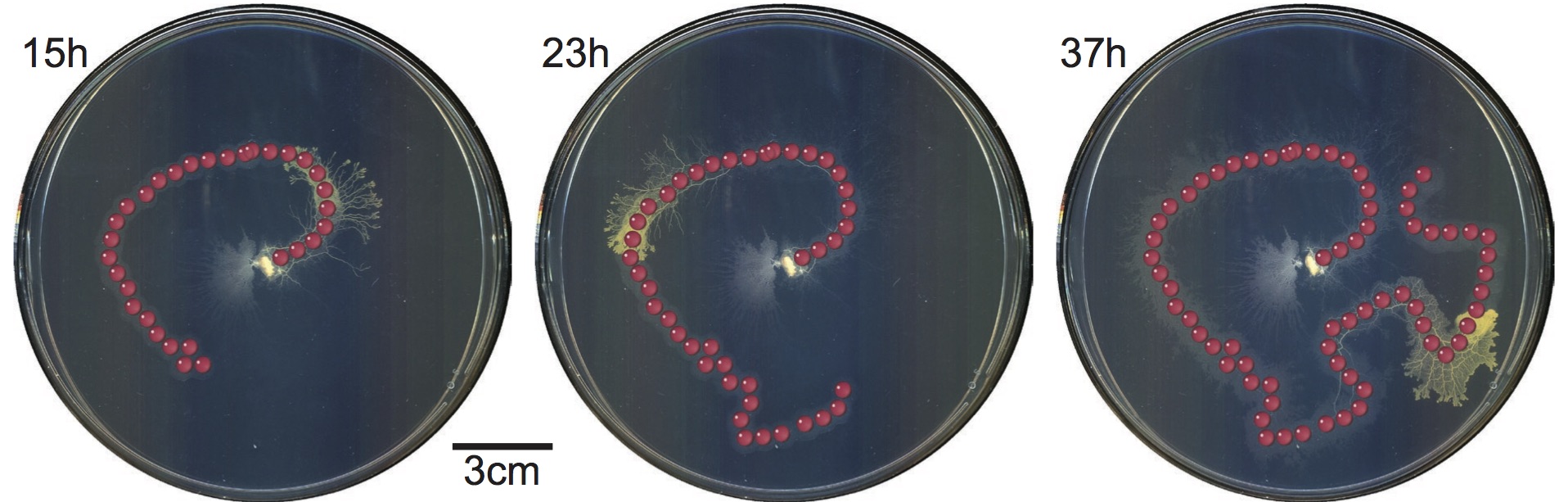
Zahid Hossain, Alice M. Chung Ingmar H. Riedel-Kruse, "Real-time and Turn-based Biology Online Experimentation" Proceedings of 12th International Conference of Remote Engineering and Virtual Instrumentation (REV), 2015.
Abstract: [PDF]
Being able to perform versatile biology experiments online would be very beneficial for research and education. Currently no such platforms exist in the life sciences. We have developed two such platforms, which are optimized for biological phenomena at the seconds and hours time scale, respectively: (1) A fully automated real- time biology lab that enables direct online interactions with phototactic microorganisms (euglena) that are long-term stably cultured in a microfluidic chip; (2) A turn-based biology lab that enables the spatio-temporal chemical stimulation of slime molds (physarum) in a petri-dish over multiple days. We compare the utility and affordance of both platforms, especially regarding use in online education.
|
|
|
|
-
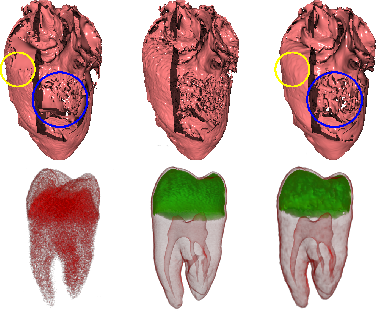
Zahid Hossain, Torsten Möller,
"Edge Aware Anisotropic Diffusion for 3D Scalar Data"
IEEE Transactions on Visualization and Computer Graphics (Proceedings Visualization / Information Visualization 2010),
vol. 16, no. 6, pp. 1375--1384, Nov.-Dec. 2010.
Abstract: [PDF] [Supplementary Materials] [IEEE Vis 2010 Slides] [Bibtex]
In this paper we present a novel anisotropic diffusion model targeted for 3D scalar field data. Our model preserves material
boundaries as well as fine tubular structures while noise is smoothed out. One of the major novelties is the use of the directional
second derivative to define material boundaries instead of the gradient magnitude for thresholding. This results in a diffusion model
that has much lower sensitivity to the diffusion parameter and smoothes material boundaries consistently compared to gradient
magnitude based techniques. We empirically analyze the stability and convergence of the proposed diffusion and demonstrate its
de-noising capabilities for both analytic and real data. We also discuss applications in the context of volume rendering.
|
|
|
|
-
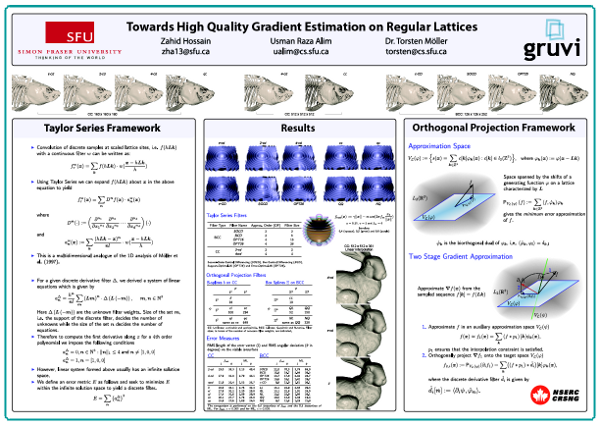
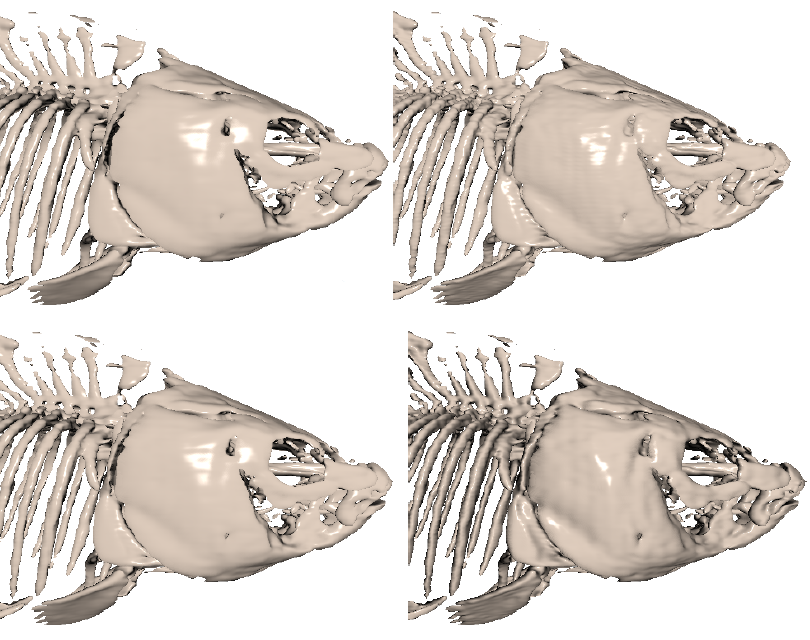
Zahid Hossain, Usman R. Alim, Torsten Möller,
"Toward High-Quality Gradient Estimation on Regular Lattices,"
IEEE Transactions on Visualization and Computer Graphics,
TVCG on 17.4 (2011): 426-439.
Abstract: [PDF] [Supplementary Materials] [Bibtex]
In this paper, we present two methods for accurate
gradient estimation from scalar field data sampled
on regular lattices. The first method is based on
the multi-dimensional Taylor series expansion of
the convolution sum and allows us to specify
design criteria such as compactness and
approximation power. The second method is based on
a Hilbert space framework and provides a minimum
error solution in the form of an orthogonal
projection operating between two approximation
spaces. Both methods lead to discrete filters
which can be combined with continuous
reconstruction kernels to yield highly accurate
estimators as compared to the current state of the
art. We demonstrate the advantages of our methods
in the context of volume rendering of data sampled
on Cartesian and Body-Centered Cubic lattices. Our
results show significant qualitative and
quantitative improvements for both synthetic and
real data, while incurring a moderate
pre-processing and storage overhead.
|
|
|
|
-
Zahid Hossain and Mohammad Towhidul Islam, Fast Ray-Triangle Intersection Test for Real-Time Applications,
in the Proceedings of the ICCIT Dhaka, Bangladesh, December, 2005.
|
|
|
|
|
Abstract: [PDF]
[Defense Slides (Keynote)]
Experimentation in life science is necessary for academia, industry and education, but many access barriers exist due to professional training, operation cost, maintenance, and safety requirements. High-throughput experimental equipment combined with cloud computing infrastructure has the potential to alleviate these access barriers through abstraction and time-sharing. Recently, a few commercial cloudlabs such as Transcriptic and Emerald Cloudlab have emerged, where highly standardized experimentation protocols are outsourced and executed semi-automatically based on the instructions provided by the users ahead of time without any means to interrupt or interact with an ongoing experiment. Therefore, a truly interactive biology cloudlab does not yet exist, whereas interaction is the key to exploration based science.
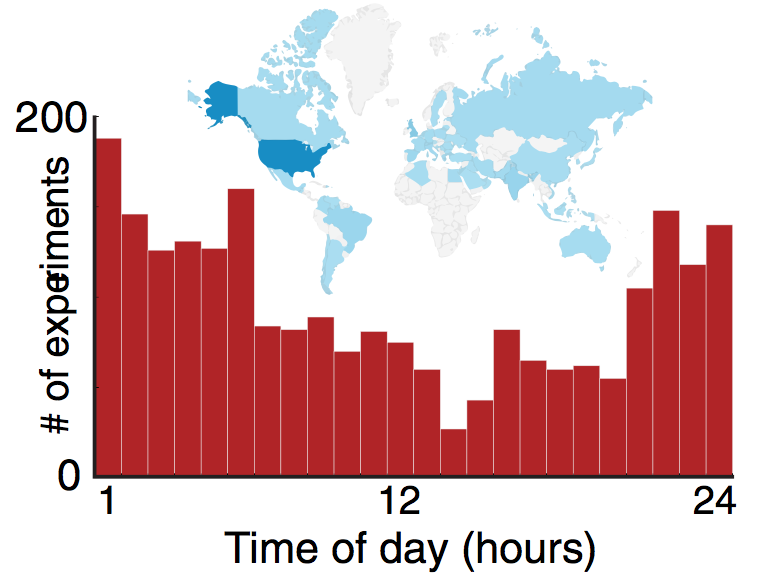
In this dissertation, we conceptualized an interactive biology cloudlab paradigm. The core of this concep- tualization is a domain specific device, Biotic Processing Unit (BPU) that hosts a biological sample and allows interactive experimentation. Biological systems are particularly difficult to handle as they, unlike physical systems, may exhibit unpredictable natural variabilities. Therefore, to provide a better Quality of Service (QoS), we proposed a general method to automatically monitor an array of backend BPUs to check the underlying biological state. We implemented two different cloudlab architectures to support 1) nonreal-time chemotaxis experimentation with a slime mold, Physarum polycephalum and 2) real-time phototaxis experimentation with a single celled micro-swimmer Euglena gracilis. In the Physarum based cloudlab, users time-shared a set of pre-allocated BPUs for experiments that would last two days. In the Euglena based cloudlab, users were scheduled in a queue using an Automatic Call Distributor (ACD) system, where experiments would last ~1 minute. We compared these two architectures to draw general design rules, and recommended future cloudlab implementations based on the time-scale of the underlying biology.
We iteratively developed our cloudlab by deploying it in various educational settings with different interaction modalities: in graduate level university classes, middle and high school classes, and eventually through an MOOC course with over 300 students. To this end, we also implemented a modeling platform along with other HCI components to facilitate visual and data analytics to enable inquiry-based learning as mandated by the National Research Council. In conclusion, we have paved the way to make complex biology experimentation accessible to a broader audience - including researchers, citizen scientists, and learners alike - at low cost and scale.
|
|
Abstract: [PDF]
[Supplementary Materials]. [Defense Slides]
We present a novel anisotropic diffusion model targeted for 3D scalar feld data. Our model
preserves material boundaries as well as fine tubular structures while noise is smoothed
out. One of the major novelties is the use of the directional second derivative to define
material boundaries instead of the gradient magnitude for thresholding. This results in a
diffusion model that has much lower sensitivity to the diffusion parameter and smoothes
material boundaries consistently compared to gradient magnitude based techniques. We
analyze the stability and convergence of the proposed diffusion and demonstrate its denoising capabilities for both analytic and real data. We also discuss applications in the
context of volume rendering.
We extend our algorithm to non-Cartesian lattices such as Body Centric Cubic (BCC).
The key to such an extension is a method to estimate derivatives reliably. Therefore, we
present a general framework to estimate derivatives on arbitrary regular lattices. With this
framework a user can design filters with compact support and specify a polynomial order of accuracy.
|
|
|
Abstract: [PDF]
[Software]
The main focus of this thesis is to solve the 2D strip-packing problem specifically for use in the
garments industries of Bangladesh. Garments industry is one of the biggest export industries of Bangladesh.
One of the routine works of all garments company is to cut shape of clothes from long strip of cloth.
The current technique for placing the shapes and cutting are both done manually which is not only time
consuming but also minimally optimized. We realized that this process of placing shapes can be automated.
However the problem itself is classified as NP-Hard problem. In this thesis we tried to solve the problem
using Genetic Algorithm keeping the orientation of the shapes fixed. This requirement of keeping the
orientation fixed is posed by the nature of the way clothes have to be cut out. In this research we
surveyed most of the works that has been done to solve similar problems and developed our new algorithm
which solves the garments industries' problem specifically. We developed software that tests our new algorithm
and presents the user with graphical output of the final placement of the shapes. Our software can also take
several input parameters which are either software specific or Genetic Algorithm specific.
|
|